
The Centro Nacional de Análisis Genómico and the VIB-KU Leuven Center for Brain & Disease Research have led this groundbreaking study, published in the prestigious journal Nature Biotechnology
The study, which is part of the Human Cell Atlas project, compiles a systematic examination of eight different single-cell accessible chromatin sequencing (scATAC-seq) technologies, revealing critical differences in data quality and performance
August 3, 2023. A study published in Nature Biotechnology has provided a comprehensive and unbiased comparison of single-cell accessible chromatin sequencing (scATAC-seq) technologies, contributing to a better understanding of gene regulation, disease mechanisms and cellular diversity. The study, titled "Benchmarking of single-cell ATAC sequencing tools," led by Holger Heyn from the Centro Nacional de Análisis Genómico (CNAG) in Barcelona (Spain) and Stein Aerts from the VIB-KU Leuven Center for Brain & Disease Research (Belgium) sheds light on the data quality achieved by each method and allows the researchers to make an informed protocol selection when considering scATAC-seq. The research is part of the Human Cell Atlas project, an open global initiative who ultimately aims to create a comprehensive map of gene regulation across all cell types, revolutionizing our knowledge of human biology.
Every cell in our body has the same DNA, but their unique functions and characteristics are driven by specific gene regulation. To unravel this complexity the research community utilizes ATAC sequencing, a modern cutting-edge technology that examines DNA accessibility. By studying the open chromatin regions of DNA, researchers can identify active regulatory elements and understand how genes are controlled in different cell types. This groundbreaking approach triggered a major revolution in epigenetics, the discipline that focuses on the changes of the chromatin and the factors that explain the DNA activity. CNAG is one of the centers in the world, specialized in Single Cell Genomics, applying these state-of-the-art protocols.
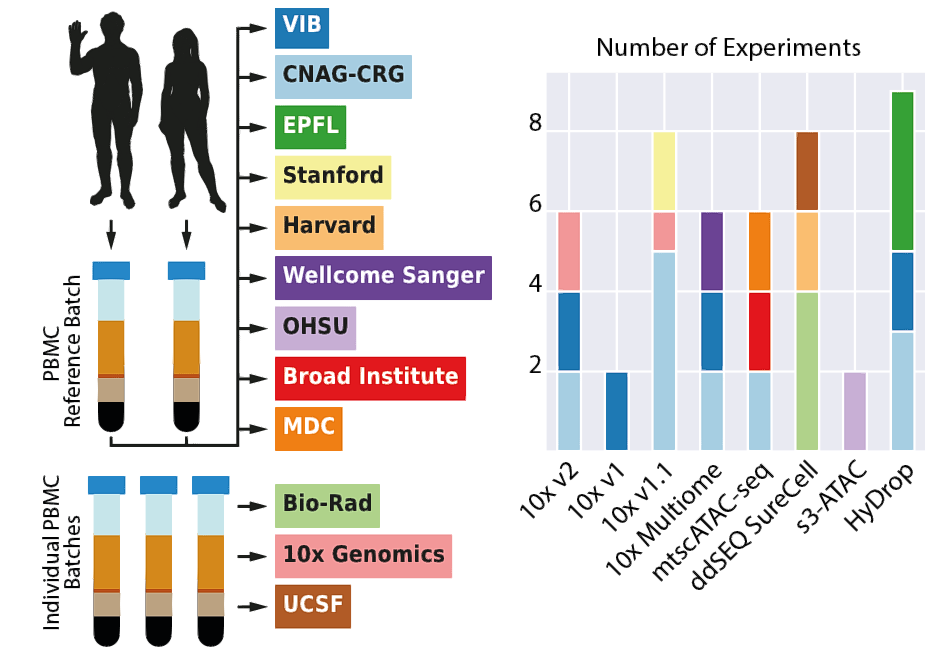
Years later, a systematic comparison of various scATAC-seq methods has still been lacking. Differences between technologies can introduce technical biases and impact the interpretation of scATAC-seq datasets. Therefore, an exhaustive benchmarking of these methods was crucial to equip researchers with the necessary knowledge for informed decision-making. For this study, research teams and commercial technology providers, including the Greenleaf and Buenrostro labs, who first published ATAC sequencing in 2013 as well as the Human Cell Atlas projects co-chairs, Aviv Regev and Sarah Teichmann, generated benchmark datasets from eight different scATAC-seq technologies (including variations of the 10x Genomics scATAC-seq protocol, 10x Multiome, Bio-Rad ddSEQ SureCell ATAC-seq, HyDrop-ATAC, and s3-ATAC). These datasets were derived from blood cells to minimize sample preparation variability, with replicates across three laboratories for most methods.
“This project is a great example of the power of collaborative science, with invaluable contributions from our partners in academia and industry. Together, we have generated datasets from 47 samples and 169,000 cells, with replicates across multiple centres, ensuring robust and fair comparison of the various technologies. We are grateful to all partner that generously contributed datasets to this unprecedented effort to derive community standards for single-cell ATAC sequencing.”, says Holger Heyn, who coordinated the study at the CNAG.

Using a new pipeline for universal mapping of scATAC-seq data (PUMATAC), created by VIB-KU Leuven Center for Brain & Disease Research, the researchers examined various parameters, including library complexity, sequencing efficiency, sensitivity, cell type annotation accuracy, and more. Striking differences were observed in the complexity of sequencing libraries and the specificity of DNA tagmentation achieved with each method. For instance, the 10x Genomics scATAC-seq (v.2) outperformed open-source methods like HyDrop and s3-ATAC by more than eight times in terms of sensitivity.
“Despite academic methods demonstrating lower performance when compared to commercial approaches, the significance of ongoing development of new technologies in laboratories worldwide is evident. Open-source and collaborative research is driving innovation for the development of the next-generation and more cost-effective ATAC sequencing methods.”, says Holger Heyn.
From the VIB-KU Leuven Center for Brain & Disease Research, Stein Aerts states: “We are entering an era where AI models combined with large, highly-resolved datasets will help us answer biology's most fundamental questions. Single-cell ATAC-seq has been a crucial component in this revolution by allowing us to accurately reconstruct cell type-specific gene regulatory networks and decipher the enhancer code embedded in the genome. The quality and cost of data remain sensitive issues. With our benchmark, we characterised the systematic variations that exist between technologies, and to set a precedent for future benchmarking and single-cell ATAC-seq data pre-processing as a whole”.
The study provides a decision-making framework for selecting appropriate scATAC-seq methods but also offers the PUMATAC pipeline for future use, ensuring seamless integration of new datasets and technologies. The researchers acknowledged the limitations of the study primarily relying on PBMC samples and emphasized the need for further exploration of performance variability in different tissues and species. This study has the potential to improve the field of single-cell epigenetic profiling by guiding method selection, improving data quality, and reducing experimental costs.
Image 1:
Wageningen University & Research
Image 2:
Study and analysis pipeline design. Summarized design of the benchmark study showing how PBMC were used as reference samples to generate 47 datasets across eight scATAC-seq techniques. Coloured rectangles indicate the different academic and industry partners that participated in the study. CNAG-CRG, Centro Nacional de Análisis Genómico; EPFL, École Polytechnique Fédérale de Lausanne; MDC, Max-Delbrück-Center for Molecular Medicine; OHSU, Oregon Health & Science University; UCSF, University of California San Francisco; VIB, Vlaams Instituut voor Biotechnologie.
Image 3:
Holger Heyn, the Single Cell Genomics Team Leader at CNAG and the coordinator of this study.
Reference article:
Florian De Rop et al., ‘Systematic benchmarking of single-cell ATAC sequencing tools’ Nature Biotechnology. August, 2023. DOI: 10.1038/s41587-023-01881-x










